| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial |
 |
| Map Type: | Genetic [ View Map Type Info ] | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Map Set Name: | Soybean - GmComposite2003 [ View Map Set Info ] | ||||||||||||||||||||||||||||||||||||||||||
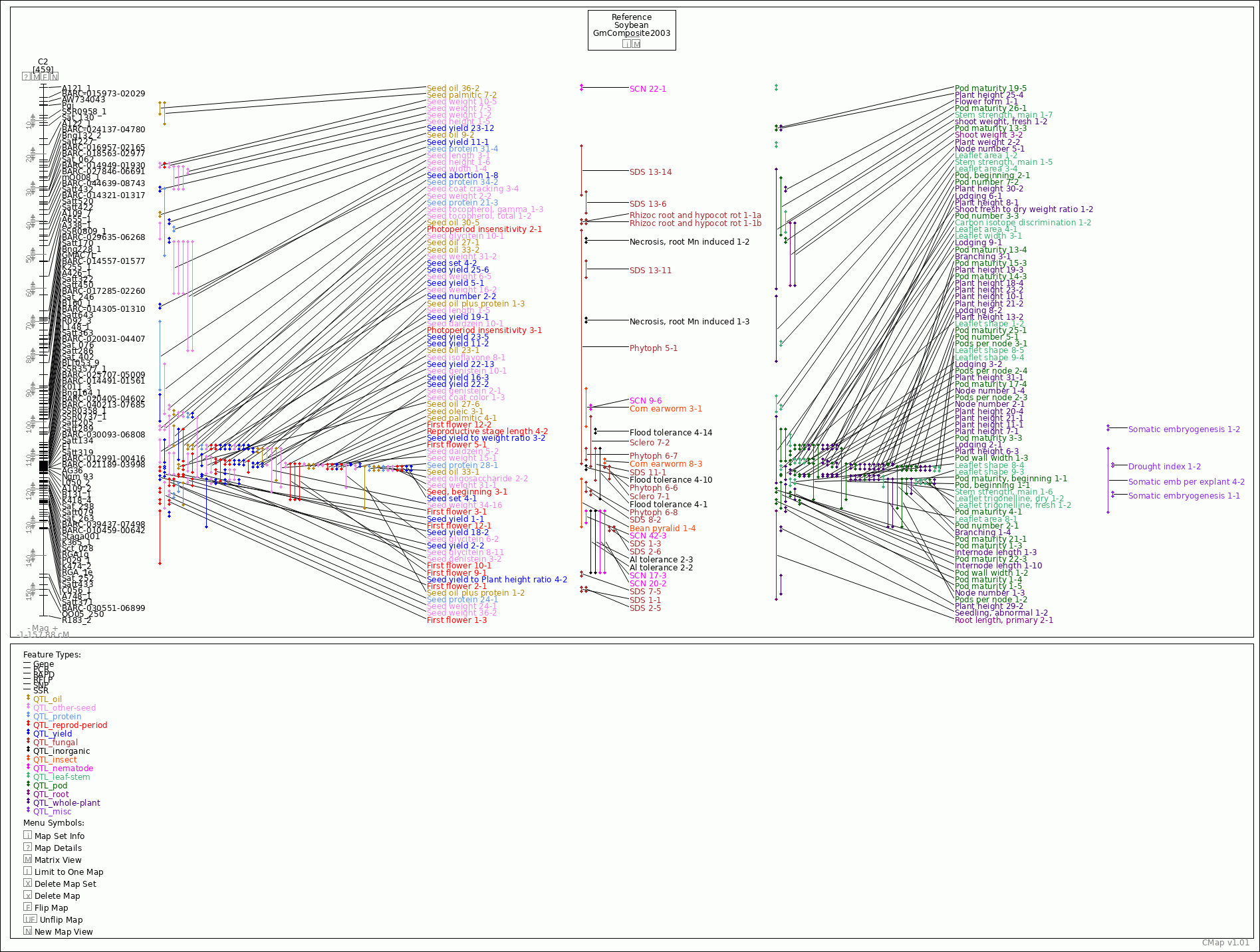
| Map Name: | C2 | ||||||||||||||||||||||||||||||||||||||||||
| Map Start: | -1 cM | ||||||||||||||||||||||||||||||||||||||||||
| Map Stop: | 157.88 cM | ||||||||||||||||||||||||||||||||||||||||||
| Features by Type: |
|
| [ Download Map Data ]
[ Download Feature Correspondence Data ] |
||||||||
| Soybean - GmComposite2003 - C2 (Click headers to resort) |
Comparative Maps | |||||||
|---|---|---|---|---|---|---|---|---|
| Feature | Type | Position (cM) | Map | Feature | Type | Position | Evidence | Actions |
| Phytoph 5-1 | QTL_fungal | 42.36 - 112.9 | No other positions | |||||
| Phytoph 6-6 | QTL_fungal | 113.38 - 113.95 | No other positions | |||||
| Phytoph 6-7 | QTL_fungal | 107.58 - 111.68 | No other positions | |||||
| Phytoph 6-8 | QTL_fungal | 117.73 - 121.26 | No other positions | |||||
| Rhizoc root and hypocot rot 1-1a | QTL_fungal | 39.3 - 41.3 | No other positions | |||||
| Rhizoc root and hypocot rot 1-1b | QTL_fungal | 39.30002 - 41.3 | No other positions | |||||
| Sclero 7-1 | QTL_fungal | 112.84 - 117.45 | No other positions | |||||
| Sclero 7-2 | QTL_fungal | 98.07 - 113.62 | No other positions | |||||
| SDS 1-1 | QTL_fungal | 149 - 151 | No other positions | |||||
| SDS 1-3 | QTL_fungal | 131 - 133 | No other positions | |||||
| SDS 11-1 | QTL_fungal | 107.58 - 117.87 | No other positions | |||||
| SDS 13-11 | QTL_fungal | 51.5 - 57.3 | No other positions | |||||
| SDS 13-14 | QTL_fungal | 17.1 - 32.74 | No other positions | |||||
| SDS 13-6 | QTL_fungal | 30.79 - 38.04 | No other positions | |||||
| SDS 2-5 | QTL_fungal | 149.00001 - 151 | No other positions | |||||
| SDS 2-6 | QTL_fungal | 131.00001 - 133 | No other positions | |||||
| SDS 7-5 | QTL_fungal | 144.48 - 146.48 | No other positions | |||||
| SDS 8-2 | QTL_fungal | 120.27 - 122.27 | No other positions | |||||
| Al tolerance 2-2 | QTL_inorganic | 126.23 - 145.47 | No other positions | |||||
| Al tolerance 2-3 | QTL_inorganic | 126.23 - 145.47 | No other positions | |||||
| Flood tolerance 4-1 | QTL_inorganic | 107.58 - 123.37 | No other positions | |||||
| Flood tolerance 4-10 | QTL_inorganic | 112.62 - 114.62 | No other positions | |||||
| Flood tolerance 4-14 | QTL_inorganic | 101.75 - 103.75 | No other positions | |||||
| Necrosis, root Mn induced 1-2 | QTL_inorganic | 44.76 - 46.76 | No other positions | |||||
| Necrosis, root Mn induced 1-3 | QTL_inorganic | 68.67 - 70.67 | No other positions | |||||